Advertisment
Erasmus virus treated by altering lung cells
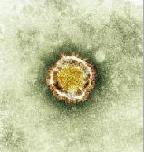
Erasmus virus resets cells’ genes and causes breathing distress and kidney failure. A new virus that causes severe breathing distress and kidney failure elicits a distinctive airway cell response to allow it to multiply. Scientists studying the Human Coronavirus-Erasmus Medical Center, which first appeared April 2012 in the Middle East, have discovered helpful details about its stronghold tactics.
Their findings predict that certain currently available compounds might treat the infection. These could act, not by killing the virus directly, but by keeping lung cells from being forced to create a hospitable environment for the virus to reproduce.
The researchers caution that their lab and computer predictions would need to be tested to see if the drugs work clinically.
The results appear in mBio, the Journal of the American Society for Microbiology.
University of Washington virologist Laurence Josset is lead author of the paper, “Cell host-response to infection with novel human coronavirus-EMC predicts potential antivirals and important differences with SARS-coronavirus.”
She conducted the research in the laboratory of senior author Michael G. Katze, UW professor of microbiology noted for pioneering systems biology approaches to host and pathogen interactions.
Eleven of the 17 reported human coronavirus EMC cases worldwide were fatal. The virus is named for the Dutch hospital that identified the specimen from a Saudi Arabia patient. So far the illness has not easily passed person to person.
The new disease agent belongs to the betacoronavirus family, as does the severe acute respiratory syndrome virus, SARS. Both viruses attack the lungs. The new virus, however, is more closely related to bat coronaviruses than to SARS. The two viruses latch onto different receptors to infect cells.
Josset, Katze and their team learned that, shortly after human coronavirus-EMC enters lung cells, it, like the SARS virus, induces changes in how the cells’ genes are regulated. But the newer virus does so sooner.
Later, and throughout infection, the human coronavirus EMC incites a massive sabotage – much greater than that of the SARS virus – of many genetic controls of protein production in lung cells grown in the laboratory.
“We found that a set of 207 genes in the lung cells was dysregulated early and permanently throughout infection with human coronavirus EMC,” Josset said. Various RNA levels were turned up or down.
The new virus appears to specifically hamper the work of several genes that enable the body to sense the presence of viruses. The scientists believe such gene re-tuning by the virus could significantly lower the ability of lung cells to mount an appropriate antiviral reaction.
While SARS and EMC activated a few similar lung cell responses for their own benefit, overall, not much overlap occurred. Each bad actor had its own modus operandi for interfering with lung cell gene activities.
University of Washington systems biologist Michael Katze (left) and virologist Laurence Josset (right) are developing rapid computational analyses of host/pathogen interactions to quickly predict possible treatments for emerging infectious diseases….click here for more information.
“These differences in host gene-expression responses in the lab-grown lung cells,” the researchers said, “might affect how each virus causes illness in an infected individual.”
At present no proven treatment exists for human coronavirus EMC. Because the virus succeeds in multiplying by hijacking cellular processes initiated in response to infection, the scientists searched for drugs that might target these cellular responses, and in so doing stop the virus from reproducing.
The researchers mentioned that this same approach is already being tested in influenza treatment. Drugs that reduce the body’s excessive inflammatory reaction to the flu virus have therapeutic benefit.
The scientists obtained a rapid, comprehensive assessment of the new coronavirus’s infective strategies by creating a global profile of how it disrupts gene transcription, the process by which DNA is copied into RNA for subsequent translation into proteins.
They analyzed this extensive data with computer programs that predict which current drugs might be re-purposed to correct the body’s virus-co-opted immune response.
The method could have widespread applications in fighting future dangerous viruses.
“Such an approach has the advantage of accelerating treatment availability, which could be crucial in case of an outbreak of an emerging pathogen,” Josset said.
Katze concurred, “Laurence and others in our group are developing new computational approaches to efficiently exploit information about the gene expression profiles induced by existing drugs and small molecules. Our goal is to quickly identify drugs that can modify specific host responses to virus infection.”
In the case of human coronavirus EMC, the approach yielded two promising possibilities. The analysis suggested that the early and sustained changes in lung cell gene regulation could be reverted by four types of kinase inhibitor and one kind of glucocorticoid.
Additional studies are necessary, the researcher said, to determine the safety, effectiveness and required dosages of these drugs in treating human coronavirus EMC.
What this study highlights, Josset said, is the advantages of fast, automated analysis of the transcriptome (all the messenger RNAs transcribed from the genome) of the infected cells.
“This method globally and efficiently characterizes the host response to emerging pathogens,” Josset said. Data on the basic properties of a new virus and its interactions with host cells can usually be collected speedily. It takes, on average, between two weeks to a month after the virus has been identified and isolated.
The gene expression data obtained from the analysis of human coronavirus EMC infection, she said, was so robust that it “provides a plethora of data to mine for further understanding and ideas to test about the new virus.”
“Because,” she added, “host response profiles also can be used to quickly identify possible treatment strategies, we anticipate that generating such profiles will become a general strategy for rapid characterization of future emerging viruses.”
Katze noted, “The emergence of new viruses, such as the H7N9 influenza virus in China, will continue to be a threat to public health. Devising new strategies to rapidly identify effective antiviral drugs is a high priority.”
###
In addition to Josset and Katze, the other researchers on the project were Vineet D. Menachery, Lisa E. Gralinski, Sudhakar Agnihothram, Boyd L. Yount, Rachel L. Graham, and Ralph S. Baric, all of the University of North Carolina at Chapel Hill, and Pavel Sova and Victoria S. Carter, both of the UW.
The project was funded in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health, contract HHSN272200800060C and grant U54AI081680.





